A COSMOS Cookbook
The following describes some procedures for handling data reduction
tasks. For detailed information on each of the programs the reader
should refer to the web pages for that program.
Reducing multislit spectra
Let's assume that you have a mask named
Mymask, and you have obtained the following set of observations
with the short camera in Nod&Shuffle orientation, using the 200
line grism
Frame
|
Exposure
|
ccd001
|
bias
|
ccd002
|
direct mask image
|
ccd003
|
spectroscopic flat
|
ccd004
|
comparison arc
|
ccd005
|
spectrum
|
ccd006
|
comparison arc
|
ccd007
|
spectrum
|
ccd008
|
comparison arc
|
ccd009
|
spectroscopic flat
|
Step 1: setting up the environment
COSMOS assumes that all the data files except the FITS image files are
in the current working directory, but that the FITS files are in
the directory pointed to by the environment variable
COSMOS_IMAGE_DIR. Everybody has their own preferences for arranging
files; one convenient way is to have all the data files for an
observing run in a directory dir, with
subdirectories n1,n2, ...
for each night's FITS files. You should cd to the
directory in
which you will keep and use the non-image files, then used setcosdir
to specify the image directory. With the above suggested
directory structure, to work with image files in the n1
subdirectory, you could either define COSMOS_IMAGE_DIR as ".",- i.e. the current
directory- and refer to image files as n1/filename,
or define COSMOS_IMAGE_DIR as "/home/joe-users/...../dir/n1",
and refer to image files as filename.
Step 2: setting up observation definition files
We need to set up an obsdef file for the spectroscopic observations. It
also doesn't hurt to first check the direct image of the mask, just to
ensure that there are no major problems. For this we need a direct
image obsdef file as well.
we invoke defineobs
and insert the following data for the direct image:
Mask:
|
Mymask
|
Dewar Offset File
|
SCdirect_NS
|
Camera:
|
SHORT f/2
|
Mode:
|
DIRECT
|
Observation Definition File
|
Mymask-direct
|
and for the spectroscopic observations, the following:
| Mask: |
Mymask
|
Dewar Offset File
|
SC200g_NS
|
Camera:
|
SHORT f/2
|
Mode:
|
SPECTROSCOPIC
|
Grism:
|
200l
|
Observation Definition
File
|
Mymask
|
Because we have not yet used align-mask on
any images, we have used the default dewar offset files, which are
located in $COSMOS_HOME/examples/dewoff.
When finished, we now have two obsdef files: Mymask-direct.obsdef,
and Mymask.obsdef.
Step 2: Aligning masks
It's usually not even necessary to look at the direct mask images, but
let's do that anyway just to make sure everything is OK (useful to do,
for example, at the beginning of a run). First, we set the parameters
in align-mask
editpar align-mask
type q to end
SEARCHBOX
10 half width of search range
MAGFACTOR
100 magnification factor for offset
vectors
LAMFILE
HNA.dat line list file
NAVER
1 number of neighboring points to
average is plot
Change parameter:
A searchbox of 10 is usually sufficient; if most apertures cannot be
found, increase it to 20. The lamfile is only needed for the
spectroscopic image alignment.
Now, run align-mask
to check and correct the alignment.
align-mask -o
Mymask-direct -f ccd002
After the first iteration, assuming that it is successful, rerun defineobs and
insert ccd002 as the name of the dewar offset file. The second
iteration should show quite good agreement.
Assuming that this worked well, we are ready to align the spectroscopic
images.
If it didn't work:
If it did not work well, something may
be very wrong. In that case we should generate a list of predicted
positions using
apertures and
overlay it in IRAF with the mosaiced image.
apertures -o
Mymask-direct -b 1 (assuming you used 1x1 binning)
In IRAF:
display8 ccd001 1
z1=0 z2=whatever
tvmark 1
Mymask.xy
A comparison of predicted and observed apertures positions should allow
you to understand the problem.
Assuming it did work:
Now we need to
align the spectroscopic mask. We only do this once, using one of the exposures, for a set of
spectroscopic exposures that we want to reduce as a group and combine
(even though flexure may result in slight shifts from one exposure to
the next.) The reason for this will be explained later. We need to
specify a line list file in the align-mask parameter file. This
list should consist of a small number of clean, well-isolated
comparison lines. In this example, we have called it HNA.dat. Having
done so:
align-mask -o
Mymask -f ccd006
Again, change the name of the dewar offset file in Mymask.obsdef
to ccd006 after the first iteration. The second iteration should look
pretty good, like this:
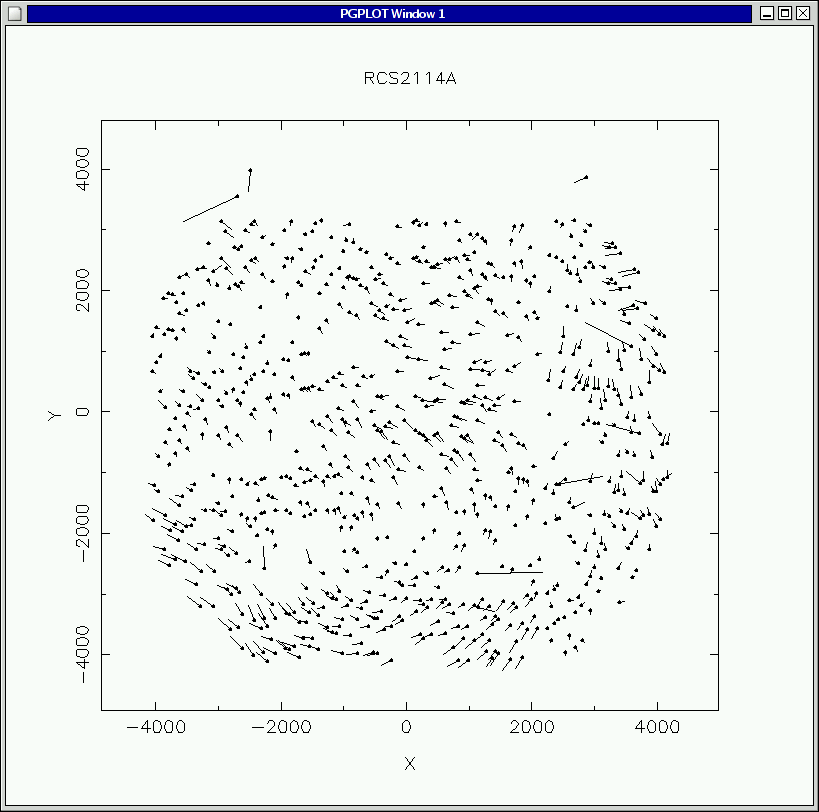
The typical residual is about a pixel, and is dominated, as is obvious,
by systematic errors. These are due to imperfectly-mapped distortions
in the IMACS optics. These small errors will be removed later when we
construct and adjust the spectral map.
Step 3: Constructing the spectral map
The spectral map is a file
which contains the information necessary to transform from CCD
coordinates to the space of wavelength vs slit position into which we
want to transform our data. The information necessary to construct a
first, approximate map (which should be good to a few pixels) is all
contained in the observation defintion file. The map is constructed
using the program map-spectra. So:
map-spectra Mymask
We will do this only once for each set of data which we wish to
combine. Because this map is approximate, we use the comparison arc
exposures to
adjust it in the most suitable way for each sicence spectral exposure,
using adjust-map. We
can use a single arc exposure, or interpolate between two. In our
example, each science exposure is bracketed by two arc exposures, so we
can do:
adjust-map
-m Mymask -f ccd004 ccd006
which will produce a new map file ccd004-6.map to
use with the science exposure ccd005, and so forth. To repeat what was stated earlier, if we
wish to later combine ccd005, and
ccd007,
with cosmic ray rejection, we should start with the same basic map
file, Mymask.map,
created from one observation definition file, Mymask.obsdef.
It's not a bad idea to check the map file output created by adjust-map, by
using spectral-map
to create a list of expected spectral line positions, then overlaying
them on the image of the arc frame.
spectral-map -m
ccd004-6 -l linelist.dat -b 1
where
linelist.dat is a file containing a comparison arc line
list.
Then, in IRAF
display8 ccd004 1
z1=0 z1=whatever
tvmark 1
ccd004-6.xy
The marked positions should align virtually perfectly with the slit
centers (to within the pixilization). If they're still off, try
repeating adjust-map:
adjust-map -m
ccd004-6 -f ccd004 ccd006
That should do it quite well.
Step 4: Preparing the frames
We now need to do bias subtraction and flat-fielding of the science
exposures, using biasflat.
Since we will use the same bias frame for all science exposures, we can
identify it in the biasflat parameter file, but we will need to specify
different flat-field frames on the command line for each science
exposure. We create spectroscopic flat fields for the science
exposures using Sflats, in
our case
Sflats -m ccd004-6
-f ccd003
will create a set of image files ccd003_flat
which we will use with ccd005.
Here's what a section of one of these frames looks like:
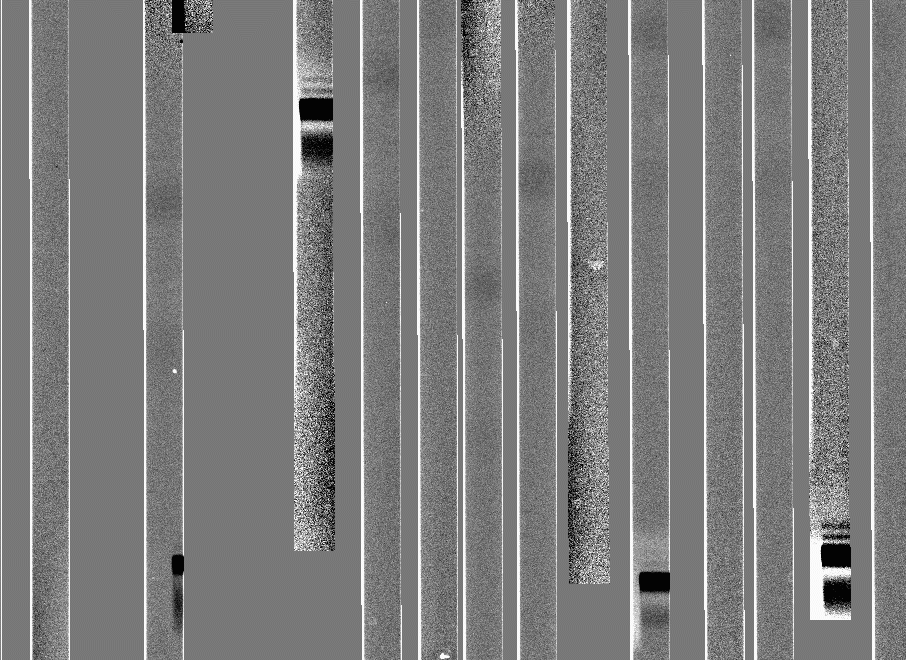
Now we are ready to biasflat ccd005:
biasflat -f
ccd003_flat ccd005
The output will be a set of images files ccd005_f.
Here's what the same section of a flattened spectrum frame looks like:
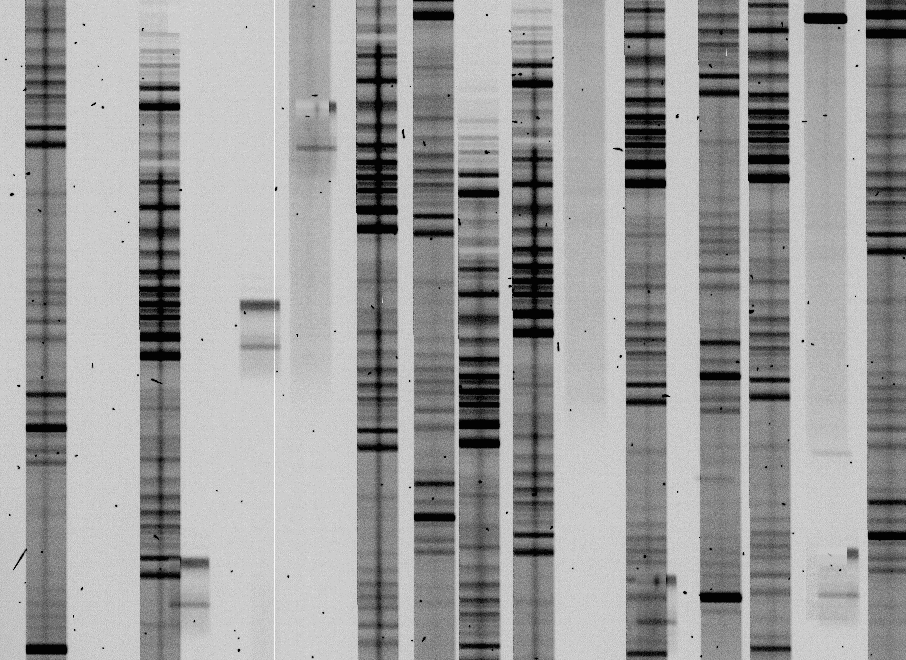
Step 5: Processing the spectra
The next step is to subtract sky using the routine subsky.
subsky -m ccd004-6
-f ccd005_f
which produces a set of image files ccd005_s,
a section of which looks like this:
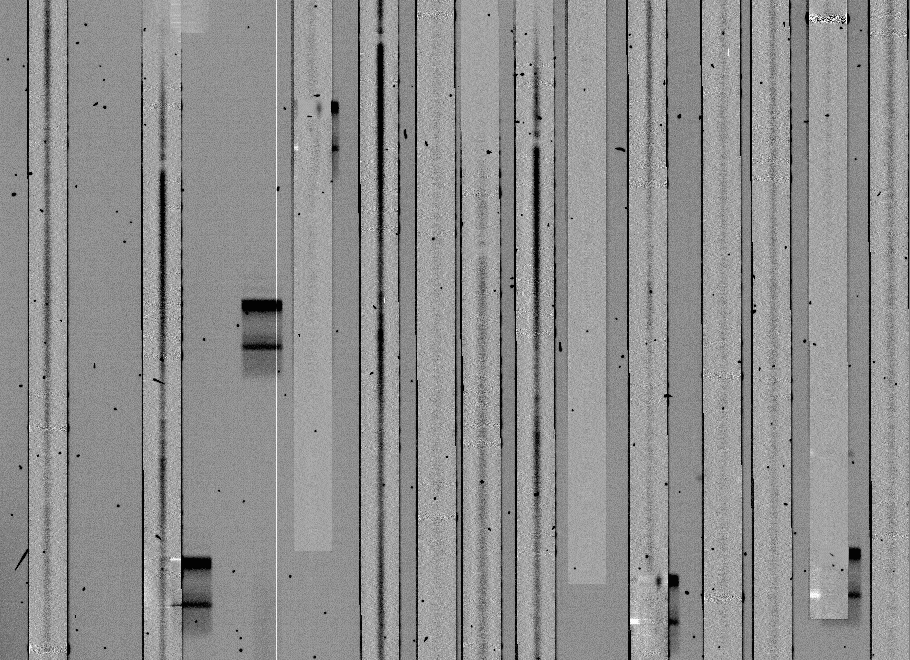
If the comparison arcs are well-matched to the object frames, the sky
subtraction should usually be this clean. If there are large residuals,
the most likely reason is a poorly-matched comparison arc (i.e.
something shifted between arc and object exposures).
We are now ready to extract the spectra You have three choices:
- Do a 2-d extraction on each object exposure, using extract-2dspec,
then use sumspec to
combine the frames with cosmic-ray rejection.
- Do a 1-d or 2-d combining+extraction+CR rejection using extract
- Using the spectrum mapping in the map file plus the sky
subtracted spectral frames, design a custom extraction procedure. See Rolling your own for more details on this.
Method 1 is the most robust at the present time, so we'll do that.
First, we extract each object exposure
extract-2dspec -m
ccd004-6 -f ccd005_s
extract-2dspec
-m ccd006-8 -f ccd007_s
Now, having turned on cosmic-ray rejection in the sumspec
parameter
file, we do:
sumspec -o
Mymask_2spec ccd005 ccd007
Note that we don't specify any spectrum type (like _2spec) for the
input files, but must for the output file.
Pipelining it
Once all this becomes routine, i.e. you know how all the programs
behave and how your data behaves, you can put everything from the adjust-map
procedures
onward into a script, using process-2spec.
Note the restrictions listed in the web page before running.
| process-2spec
|
|
|
|
| Spectrum
set: |
Mymask_night1
|
| Associated
obsdef file: |
Mymask
|
|
|
| Science
frame # |
005
|
|
Bias
frame: |
bias
|
|
Flat
frames: |
003
|
|
Comparison
arcs: |
004
006
|
|
|
| Science
frame # |
007
|
|
Bias
frame: |
bias
|
|
Flat
frames: |
009
|
|
Comparison
arcs: |
006
008
|
|
|
| Science
frame # |
|
Spectrum reduction
makefile Mymask_night1.make created
The script Mymask_night1.make
can
be executed using the standard unix command make.
make
-f Mymask_night1.make



